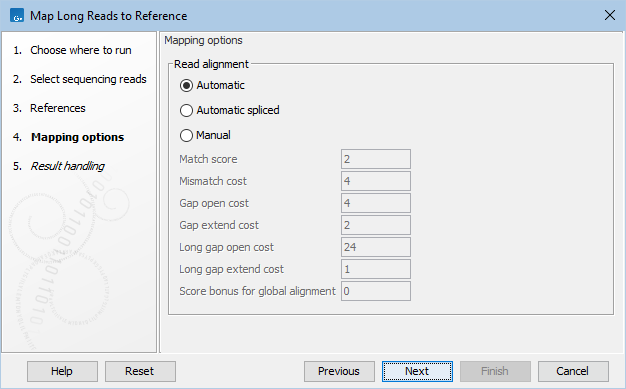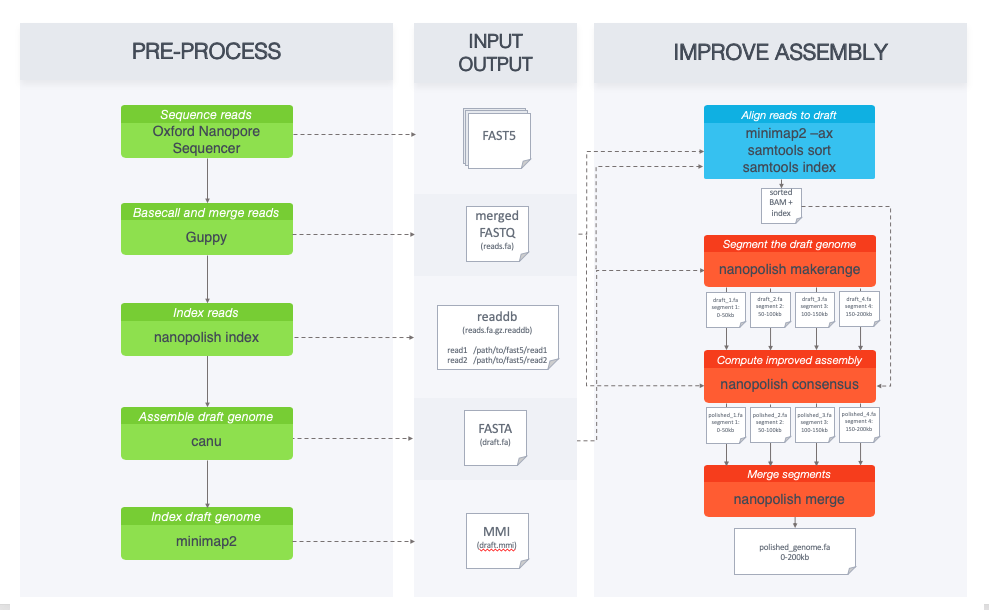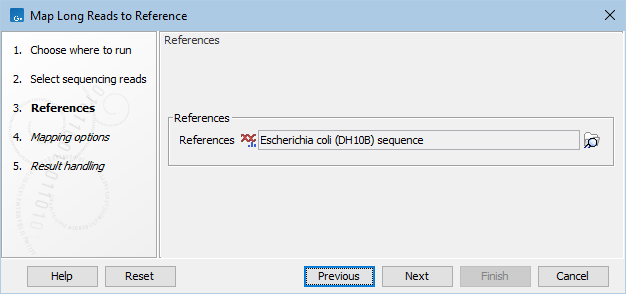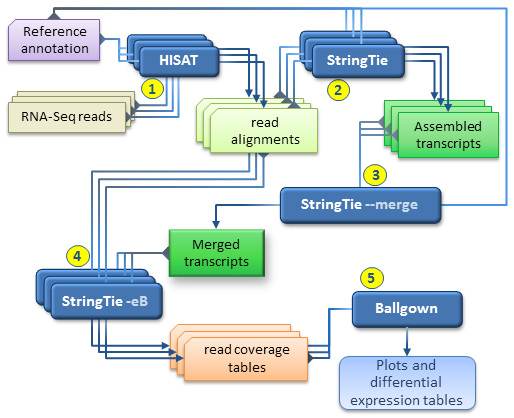
Frontiers | A Novel SARS-CoV-2 Viral Sequence Bioinformatic Pipeline Has Found Genetic Evidence That the Viral 3′ Untranslated Region (UTR) Is Evolving and Generating Increased Viral Diversity

Minimap2 workflow depicting its three key modules: (i) seeding, (ii)... | Download Scientific Diagram

Microorganisms | Free Full-Text | Nanopore Sequencing Is a Credible Alternative to Recover Complete Genomes of Geminiviruses

Accelerated identification of disease-causing variants with ultra-rapid nanopore genome sequencing | Nature Biotechnology

Minimap2 workflow depicting its three key modules: (i) seeding, (ii)... | Download Scientific Diagram

Introduction to the principles and methods underlying the recovery of metagenome‐assembled genomes from metagenomic data - Goussarov - 2022 - MicrobiologyOpen - Wiley Online Library